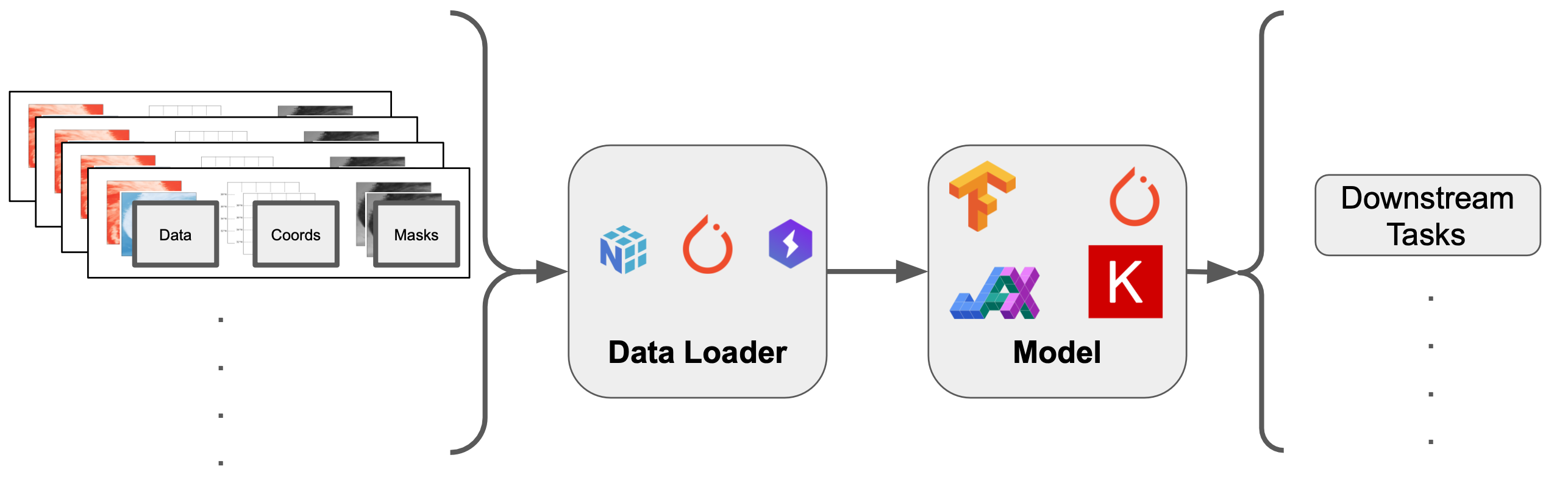
ML-Ready Data¶
In this tutorial, we will go over some of the basics to create dataloaders.
In [2]:
Copied!
import autoroot
import os
import xarray as xr
import matplotlib.pyplot as plt
from xrpatcher import XRDAPatcher
from torch.utils.data import Dataset, DataLoader, ConcatDataset
import numpy as np
import itertools
from dotenv import load_dotenv
from rs_tools._src.utils.io import get_list_filenames
xr.set_options(
keep_attrs=True,
display_expand_data=False,
display_expand_coords=False,
display_expand_data_vars=False,
display_expand_indexes=False
)
np.set_printoptions(threshold=10, edgeitems=2)
save_dir = os.getenv("ITI_DATA_SAVEDIR")
import autoroot
import os
import xarray as xr
import matplotlib.pyplot as plt
from xrpatcher import XRDAPatcher
from torch.utils.data import Dataset, DataLoader, ConcatDataset
import numpy as np
import itertools
from dotenv import load_dotenv
from rs_tools._src.utils.io import get_list_filenames
xr.set_options(
keep_attrs=True,
display_expand_data=False,
display_expand_coords=False,
display_expand_data_vars=False,
display_expand_indexes=False
)
np.set_printoptions(threshold=10, edgeitems=2)
save_dir = os.getenv("ITI_DATA_SAVEDIR")
ML-Ready Datasets¶
In [3]:
Copied!
list_of_files = get_list_filenames(f"{save_dir}/aqua/analysis", ".nc")
len(list_of_files)
list_of_files = get_list_filenames(f"{save_dir}/aqua/analysis", ".nc")
len(list_of_files)
Out[3]:
96
In [4]:
Copied!
ds = xr.open_dataset(list_of_files[0], engine="netcdf4")
ds
ds = xr.open_dataset(list_of_files[0], engine="netcdf4")
ds
Out[4]:
<xarray.Dataset> Size: 169kB Dimensions: (y: 32, x: 32, band: 38, time: 1, band_wavelength: 38) Coordinates: (6) Dimensions without coordinates: y, x Data variables: (1)
PyTorch Integration¶
In [5]:
Copied!
from rs_tools._src.utils.io import get_list_filenames
from rs_tools._src.datamodule.utils import load_nc_file
from rs_tools._src.datamodule.editor import StackDictEditor, CoordNormEditor
from toolz import compose_left
from rs_tools._src.utils.io import get_list_filenames
from rs_tools._src.datamodule.utils import load_nc_file
from rs_tools._src.datamodule.editor import StackDictEditor, CoordNormEditor
from toolz import compose_left
We will create a very simple demo dataloader
In [6]:
Copied!
from torch.utils.data import Dataset, DataLoader
from typing import Optional, Callable
class NCDataReader(Dataset):
def __init__(self, data_dir: str, ext: str=".nc", transforms: Optional[Callable]=None):
self.data_dir = data_dir
self.data_filenames = get_list_filenames(data_dir, ext)
self.transforms = transforms
def __getitem__(self, ind) -> np.ndarray:
nc_path = self.data_filenames[ind]
x = load_nc_file(nc_path)
if self.transforms is not None:
x = self.transforms(x)
return x
def __len__(self):
return len(self.data_filenames)
from torch.utils.data import Dataset, DataLoader
from typing import Optional, Callable
class NCDataReader(Dataset):
def __init__(self, data_dir: str, ext: str=".nc", transforms: Optional[Callable]=None):
self.data_dir = data_dir
self.data_filenames = get_list_filenames(data_dir, ext)
self.transforms = transforms
def __getitem__(self, ind) -> np.ndarray:
nc_path = self.data_filenames[ind]
x = load_nc_file(nc_path)
if self.transforms is not None:
x = self.transforms(x)
return x
def __len__(self):
return len(self.data_filenames)
In [7]:
Copied!
ds = NCDataReader(f"{save_dir}/aqua/analysis")
dl = DataLoader(ds, batch_size=1)
ds = NCDataReader(f"{save_dir}/aqua/analysis")
dl = DataLoader(ds, batch_size=1)
In [8]:
Copied!
out = next(iter(dl))
out = next(iter(dl))
In [9]:
Copied!
list(out.keys())
list(out.keys())
Out[9]:
['data', 'wavelengths', 'coords', 'cloud_mask']
In [10]:
Copied!
out["data"].shape, out["coords"].shape
out["data"].shape, out["coords"].shape
Out[10]:
(torch.Size([1, 38, 32, 32]), torch.Size([1, 2, 32, 32]))
Transforms/Editors¶
We can also use custom transformations within the dataset (just like standard PyTorch) to transform our dataset
In [11]:
Copied!
transforms = compose_left(
CoordNormEditor(),
StackDictEditor(),
)
transforms = compose_left(
CoordNormEditor(),
StackDictEditor(),
)
In [ ]:
Copied!
# initialize dataset with transforms
ds = NCDataReader(f"{save_dir}/aqua/analysis", transforms=transforms)
# initialize dataloader
dl = DataLoader(ds, batch_size=1)
# do one iteration
out = next(iter(dl))
# inspect a batch
out.shape
# initialize dataset with transforms
ds = NCDataReader(f"{save_dir}/aqua/analysis", transforms=transforms)
# initialize dataloader
dl = DataLoader(ds, batch_size=1)
# do one iteration
out = next(iter(dl))
# inspect a batch
out.shape
In [ ]:
Copied!